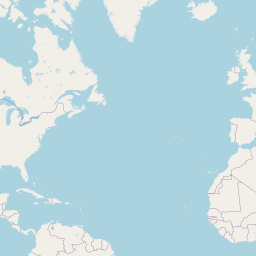
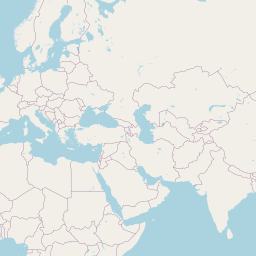
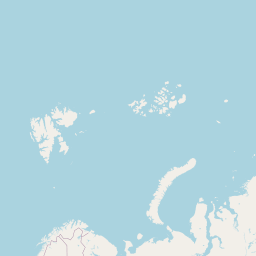
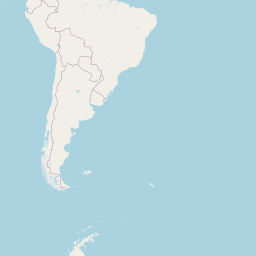
Code
search_date <- "2023-01-06"
combined_data <- read_rds(here("others", "data", "raw_data", paste0(search_date, "_data.rds")))
project_crs <- "EPSG:4326"
rodent_locations <- combined_data$rodent %>%
select(genus, species, latitude, longitude, number) %>%
mutate(number = as.numeric(number)) %>%
drop_na(number) %>%
st_as_sf(coords = c("longitude", "latitude"), crs = project_crs) %>%
group_by(geometry, species) %>%
mutate(number = sum(number)) %>%
ungroup() %>%
distinct() %>%
rowwise() %>%
mutate(labels = HTML(paste0("Species: ", species, "<br>", "Number detected: ", number)))
pal <- colorFactor(diverging_hcl(n = length(sort(unique(rodent_locations$genus))), palette = "Berlin"),
domain = sort(unique(rodent_locations$genus)))
leaflet(data = rodent_locations) %>%
addTiles() %>%
addCircleMarkers(radius = ~ifelse(sqrt(number) == 0, 1, sqrt(number)),
color = ~pal(genus),
stroke = FALSE,
fillOpacity = 0.8,
clusterOptions = markerClusterOptions(spiderfyOnMaxZoom = TRUE,
spiderLegPolylineOptions = list(length = 6),
spiderfyDistanceMultiplier = 2),
label = ~labels,
popup = ~labels) %>%
addLegend("bottomright", pal = pal, values = ~genus, title = "Host genus", opacity = 0.8)An interactive map displaying the location of detected small species in included studies. Selecting points will expand the number of data points at those coordinates. Point colour indicates small mammal genus with size of the point varying by the number of individuals detected. Hovering over a point or selecting it will show the species name, the number detected and the time period surveyed at the location. Data is currently shown for 9 studies.